Revista Mexicana de Ingeniería Química, Vol. 18, No. 3 (2019), Bio455
Investigation of novel Laccase producing fungal species and its substrate specificity by wet and dry lab
|
M. Aftab, A. Tahir, T. Asim, A. Nawaz, Y. Saleem
https://doi.org/10.24275/rmiq/Bio455
Abstract
 |
|
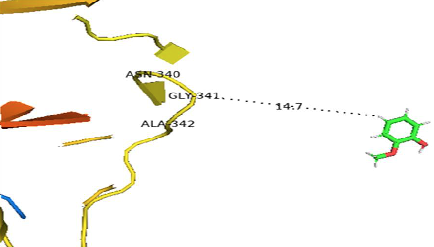
Fungal laccase has potential to degrade organic pollutants of waste water in the presence of redox mediators. Choice of better enzyme-redox mediator system can enhance the degradation of recalcitrant aromatic compounds. Present study aimed at the screening and identification of laccase producing fungal strain. Evaluate its substrate specificity to understand the interaction between the enzyme and different substrates. A total of 30 fungal strains were isolated from air, soil and decaying wood of mangifera indica and screened for their ability to produce laccase. The enzyme was produced on sawdust, a cheap substrate under static condition. Specie identification of the most potent strain was carried out by DNA barcoding. Tree building tool was used to get the phylogenetic relationship. Best enzyme substrate complex was determined by combining wet and dry lab investigations using different substrates. Predicted protein model was generated and used for docking analysis with the mediators. Only 15 strains exhibited laccase producing capability and the strain MFw3 showed highest enzyme activity 297±1 U/ml/min. Phylogenetic tree analysis showed the novelity of Aspergillus flavus MFw3 (GenBank accession number: MF167358). Bioinformatics tools were used to get the best enzyme/substrate complex and ABTS was found best substrate with minimum dock score (30.2). Our results showed that MFw3 is a novel strain and has great potential for laccase production by using ABTS as a substrate.
Keywords: Laccase, sequence analysis, ABTS, docking.
|
|
 |

